Finding bottlenecks with cProfile
After assessing the correctness and timing the execution time of the program, we are ready to identify the parts of the code that need to be tuned for performance. Those parts are typically quite small compared to the size of the program.
Two profiling modules are available through the Python standard library:
- The
profilemodule: This module is written in pure Python and adds a significant overhead to the program execution. Its presence in the standard library is because of its vast platform support and because it is easier to extend. - The
cProfilemodule: This is the main profiling module, with an interface equivalent toprofile. It is written in C, has a small overhead, and is suitable as a general purpose profiler.
The cProfile module can be used in three different ways:
- From the command line
- As a Python module
- With IPython
cProfile does not require any change in the source code and can be executed directly on an existing Python script or function. You can use cProfile from the command line in this way:
$ python -m cProfile simul.pyThis will print a long output containing several profiling metrics of all of the functions called in the application. You can use the -s option to sort the output by a specific metric. In the following snippet ,the output is sorted by the tottime metric, which will be described here:
$ python -m cProfile -s tottime simul.pyThe data produced by cProfile can be saved in an output file by passing the -o option. The format that cProfile uses is readable by the stats module and other tools. The usage of the -ooption is as follows:
$ python -m cProfile -o prof.out simul.pyThe usage of cProfile as a Python module requires invoking the cProfile.run function in the following way:
from simul import benchmark
import cProfile
cProfile.run("benchmark()")You can also wrap a section of code between method calls of a cProfile.Profile object, as shown:
from simul import benchmark
import cProfile
pr = cProfile.Profile()
pr.enable()
benchmark()
pr.disable()
pr.print_stats()cProfile can also be used interactively with IPython. The %prun magic command lets you profile an individual function call, as illustrated:
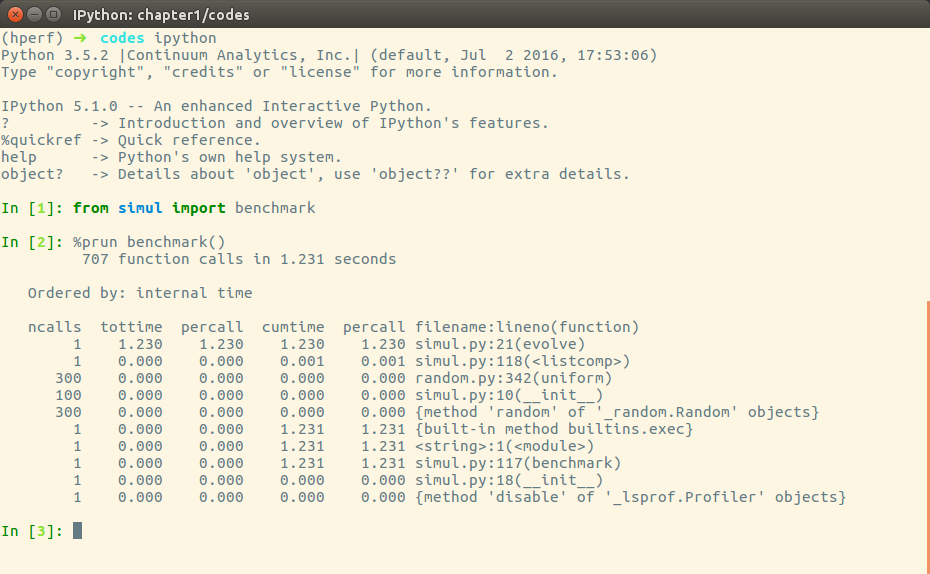
The cProfile output is divided into five columns:
ncalls: The number of times the function was called.tottime: The total time spent in the function without taking into account the calls to other functions.cumtime: The time in the function including other function calls.percall: The time spent for a single call of the function--it can be obtained by dividing the total or cumulative time by the number of calls.filename:lineno: The filename and corresponding line numbers. This information is not available when calling C extensions modules.
The most important metric is tottime, the actual time spent in the function body excluding subcalls, which tell us exactly where the bottleneck is.
Unsurprisingly, the largest portion of time is spent in the evolve function. We can imagine that the loop is the section of the code that needs performance tuning.cProfile only provides information at the function level and does not tell us which specific statements are responsible for the bottleneck. Fortunately, as we will see in the next section, the line_profiler tool is capable of providing line-by-line information of the time spent in the function.
Analyzing the cProfile text output can be daunting for big programs with a lot of calls and subcalls. Some visual tools aid the task by improving navigation with an interactive, graphical interface.
KCachegrind is a Graphical User Interface (GUI) useful to analyze the profiling output emitted by cProfile.
Note
KCachegrind is available in the Ubuntu 16.04 official repositories. The Qt port, QCacheGrind, can be downloaded for Windows from http://sourceforge.net/projects/qcachegrindwin/. Mac users can compile QCacheGrind using Mac Ports (http://www.macports.org/) by following the instructions present in the blog post at http://blogs.perl.org/users/rurban/2013/04/install-kachegrind-on-macosx-with-ports.html.
KCachegrind can't directly read the output files produced by cProfile. Luckily, the pyprof2calltree third-party Python module is able to convert the cProfile output file into a format readable by KCachegrind.
Note
You can install pyprof2calltree from the Python Package Index using the command pip install pyprof2calltree.
To best show the KCachegrind features, we will use another example with a more diversified structure. We define a recursive function, factorial, and two other functions that use factorial, named taylor_exp and taylor_sin. They represent the polynomial coefficients of the Taylor approximations of exp(x) and sin(x):
def factorial(n):
if n == 0:
return 1.0
else:
return n * factorial(n-1)
def taylor_exp(n):
return [1.0/factorial(i) for i in range(n)]
def taylor_sin(n):
res = []
for i in range(n):
if i % 2 == 1:
res.append((-1)**((i-1)/2)/float(factorial(i)))
else:
res.append(0.0)
return res
def benchmark():
taylor_exp(500)
taylor_sin(500)
if __name__ == '__main__':
benchmark()To access profile information, we first need to generate the cProfile output file:
$ python -m cProfile -o prof.out taylor.pyThen, we can convert the output file with pyprof2calltree and launch KCachegrind:
$ pyprof2calltree -i prof.out -o prof.calltree $ kcachegrind prof.calltree # or qcachegrind prof.calltree
The output is shown in the following screenshot:
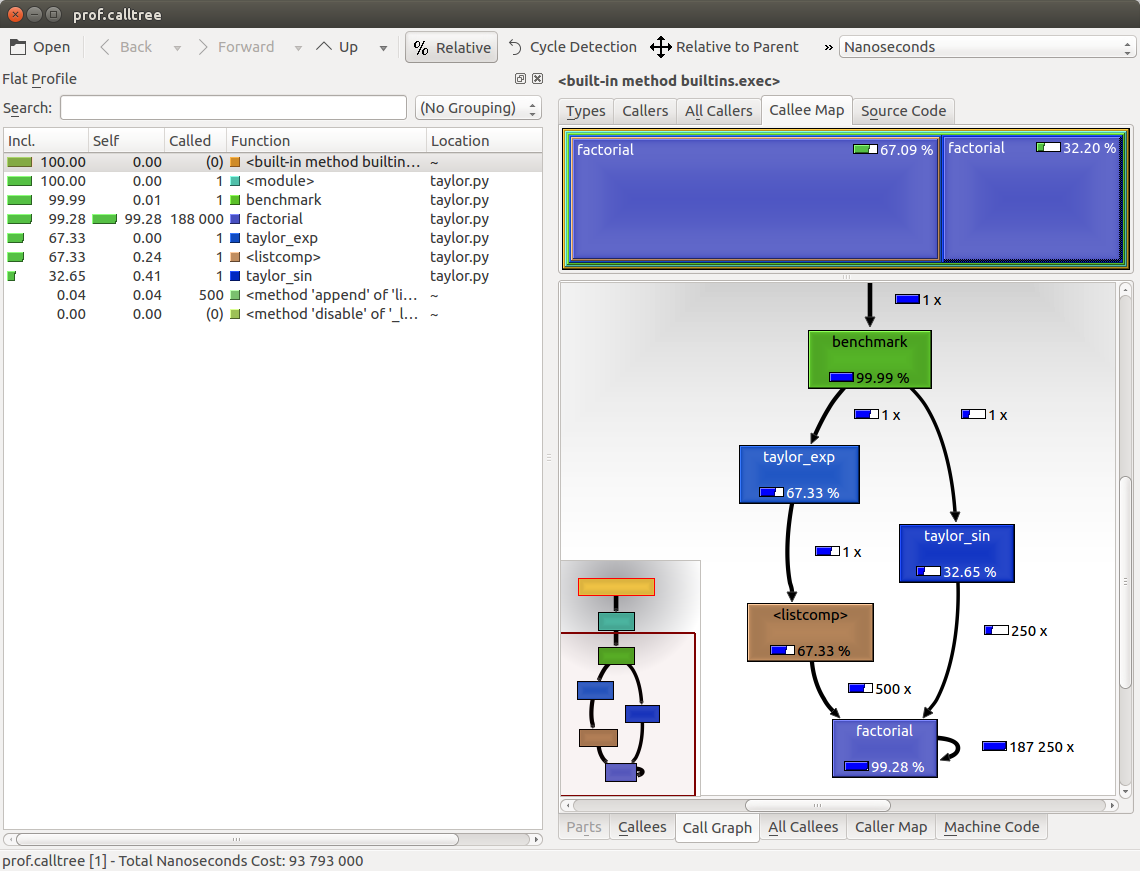
The preceding screenshot shows the KCachegrind user interface. On the left, we have an output fairly similar to cProfile. The actual column names are slightly different: Incl. translates to cProfile module's cumtime and Self translates to tottime. The values are given in percentages by clicking on the Relative button on the menu bar. By clicking on the column headers, you can sort them by the corresponding property.
On the top right, a click on the Callee Map tab will display a diagram of the function costs. In the diagram, the time percentage spent by the function is proportional to the area of the rectangle. Rectangles can contain sub-rectangles that represent subcalls to other functions. In this case, we can easily see that there are two rectangles for the factorial function. The one on the left corresponds to the calls made by taylor_exp and the one on the right to the calls made by taylor_sin.
On the bottom right, you can display another diagram, the call graph, by clicking on the Call Graph tab. A call graph is a graphical representation of the calling relationship between the functions; each square represents a function and the arrows imply a calling relationship. For example, taylor_exp calls factorial 500 times, and taylor_sin calls factorial 250 times. KCachegrind also detects recursive calls: factorial calls itself 187250 times.
You can navigate to the Call Graph or the Caller Map tab by double-clicking on the rectangles; the interface will update accordingly, showing that the timing properties are relative to the selected function. For example, double-clicking on taylor_exp will cause the graph to change, showing only the contribution of taylor_exp to the total cost.
Note
Gprof2Dot (https://github.com/jrfonseca/gprof2dot) is another popular tool used to produce call graphs. Starting from output files produced by one of the supported profilers, it will generate a .dot diagram representing the call graph.

































































