Introducing SparkSession
In Spark 2.0, SparkSession represents a unified point for manipulating data in Spark. It minimizes the number of different contexts a developer has to use while working with Spark. SparkSession replaces multiple context objects, such as the SparkContext, SQLContext, and HiveContext. These contexts are now encapsulated within the SparkSession object.
In Spark programs, we use the builder design pattern to instantiate a SparkSession object. However, in the REPL environment (that is, in a Spark shell session), the SparkSession is automatically created and made available to you via an instance object called Spark.
At this time, start the Spark shell on your computer to interactively execute the code snippets in this section. As the shell starts up, you will notice a bunch of messages appearing on your screen, as shown in the following figure. You should see messages displaying the availability of a SparkSession object (as Spark), Spark version as 2.2.0, Scala version as 2.11.8, and the Java version as 1.8.x.
The SparkSession object can be used to configure Spark's runtime config properties. For example, the two main resources that Spark and Yarn manage are the CPU the memory. If you want to set the number of cores and the heap size for the Spark executor, then you can do that by setting the spark.executor.cores and the spark.executor.memory properties, respectively. In this example, we set these runtime properties to 2 cores and 4 GB, respectively, as shown:
scala> spark.conf.set("spark.executor.cores", "2")
scala> spark.conf.set("spark.executor.memory", "4g")The SparkSession object can be used to read data from various sources, such as CSV, JSON, JDBC, stream, and so on. In addition, it can be used to execute SQL statements, register User Defined Functions (UDFs), and work with Datasets and DataFrames. The following illustrates some of these basic operations in Spark.
For this example, we use the breast cancer database created by Dr. William H. Wolberg, University of Wisconsin Hospitals, Madison. You can download the original Dataset from https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Original). Each row in the dataset contains the sample number, nine cytological characteristics of breast fine needle aspirates graded 1 to 10, and the class label , benign (2) or malignant (4).
First, we define a schema for the records in our file. The field descriptions are available at the Dataset's download site.
scala> import org.apache.spark.sql.types._
scala> val recordSchema = new StructType().add("sample", "long").add("cThick", "integer").add("uCSize", "integer").add("uCShape", "integer").add("mAdhes", "integer").add("sECSize", "integer").add("bNuc", "integer").add("bChrom", "integer").add("nNuc", "integer").add("mitosis", "integer").add("clas", "integer")
Next, we create a DataFrame from our input CSV file using the schema defined in the preceding step:
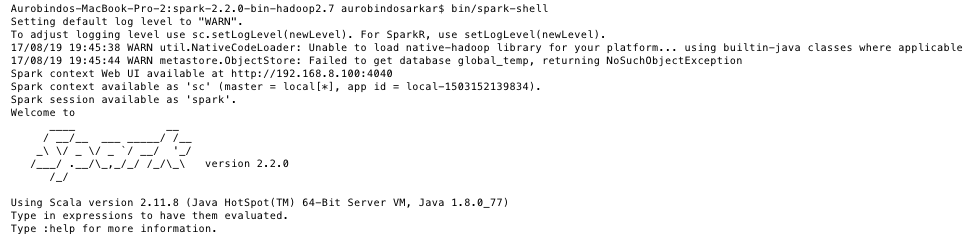
val df = spark.read.format("csv").option("header", false).schema(recordSchema).load("file:///Users/aurobindosarkar/Downloads/breast-cancer-wisconsin.data")The newly created DataFrame can be displayed using the show() method:
The DataFrame can be registered as a SQL temporary view using the createOrReplaceTempView() method. This allows applications to run SQL queries using the sql function of the SparkSession object and return the results as a DataFrame.
Next, we create a temporary view for the DataFrame and a simple SQL statement against it:
scala> df.createOrReplaceTempView("cancerTable")
scala> val sqlDF = spark.sql("SELECT sample, bNuc from cancerTable") The contents of results DataFrame are displayed using the show() method:

In the next code snippet, we show you the statements for creating a Spark Dataset using a case class and the toDS() method. Then, we define a UDF to convert the clas column, currently containing 2's and 4's to 0's and 1's respectively. We register the UDF using the SparkSession object and it in a SQL statement:
scala> case class CancerClass(sample: Long, cThick: Int, uCSize: Int, uCShape: Int, mAdhes: Int, sECSize: Int, bNuc: Int, bChrom: Int, nNuc: Int, mitosis: Int, clas: Int)
scala> val cancerDS = spark.sparkContext.textFile("file:///Users/aurobindosarkar/Documents/SparkBook/data/breast-cancer-wisconsin.data").map(_.split(",")).map(attributes => CancerClass(attributes(0).trim.toLong, attributes(1).trim.toInt, attributes(2).trim.toInt, attributes(3).trim.toInt, attributes(4).trim.toInt, attributes(5).trim.toInt, attributes(6).trim.toInt, attributes(7).trim.toInt, attributes(8).trim.toInt, attributes(9).trim.toInt, attributes(10).trim.toInt)).toDS()
scala> def binarize(s: Int): Int = s match {case 2 => 0 case 4 => 1 }
scala> spark.udf.register("udfValueToCategory", (arg: Int) => binarize(arg))
scala> val sqlUDF = spark.sql("SELECT *, udfValueToCategory(clas) from cancerTable")
scala> sqlUDF.show()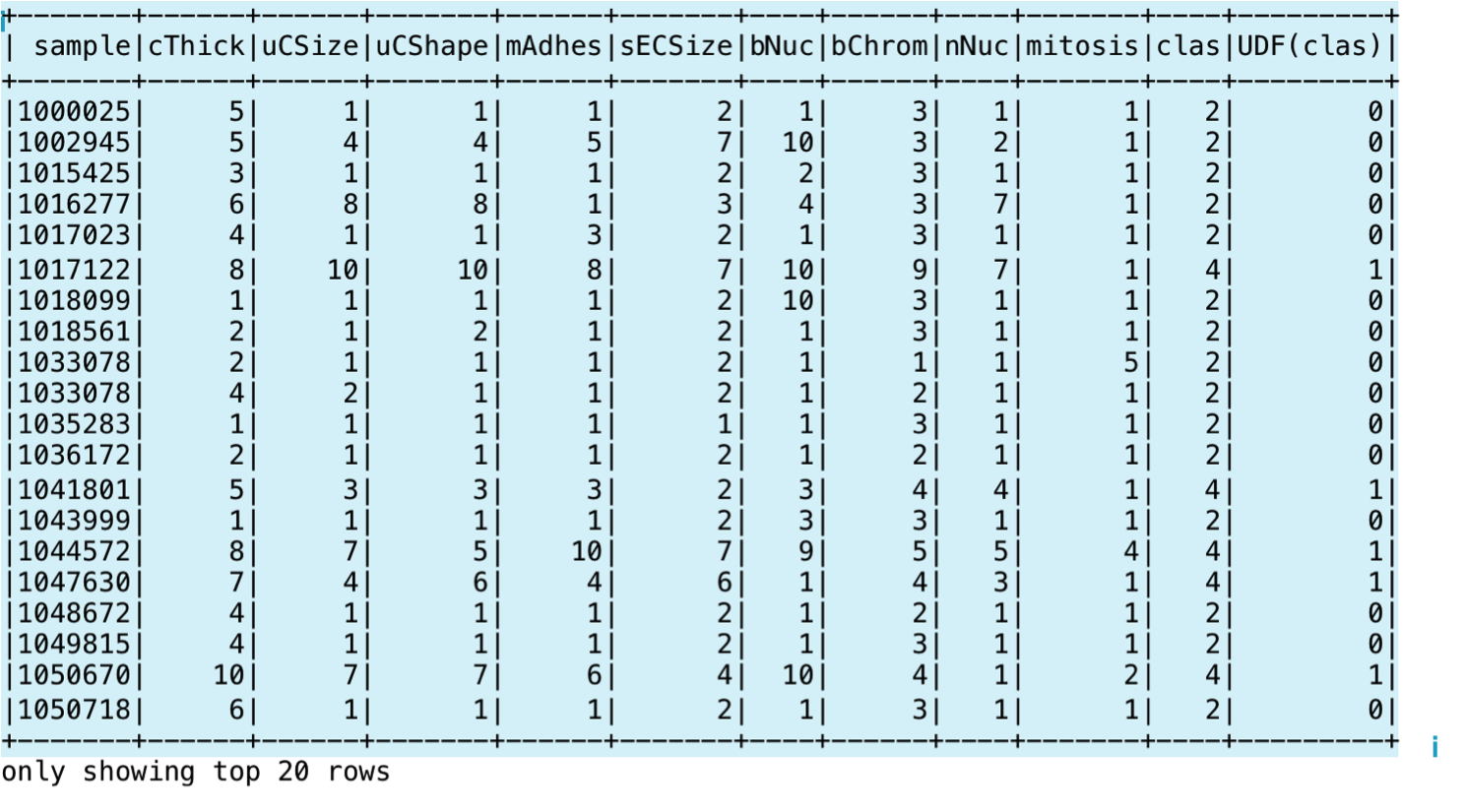
SparkSession exposes methods (via the catalog attribute) of accessing the underlying metadata, such as the available databases and tables, registered UDFs, temporary views, and so on. Additionally, we can also cache tables, drop temporary views, and clear the cache. Some of these statements and corresponding output are shown here:
scala> spark.catalog.currentDatabase
res5: String = default
scala> spark.catalog.isCached("cancerTable")
res6: Boolean = false
scala> spark.catalog.cacheTable("cancerTable")
scala> spark.catalog.isCached("cancerTable")
res8: Boolean = true
scala> spark.catalog.clearCache
scala> spark.catalog.isCached("cancerTable")
res10: Boolean = false
scala> spark.catalog.listDatabases.show()
can also use the take method to display a specific number of records in the DataFrame:
scala> spark.catalog.listDatabases.take(1) res13: Array[org.apache.spark.sql.catalog.Database] = Array(Database[name='default', description='Default Hive database', path='file:/Users/aurobindosarkar/Downloads/spark-2.2.0-bin-hadoop2.7/spark-warehouse']) scala> spark.catalog.listTables.show()

We can drop the temp table we created earlier with the following statement:
scala> spark.catalog.dropTempView("cancerTable")
scala> spark.catalog.listTables.show()
In the next few sections, we will describe RDDs, DataFrames, and Dataset constructs in more detail.