Loading data from databases
The great advantage of using a dedicated database backend instead of loading data from the disk on demand is that databases provide:
- Faster access to the whole or selected parts of large tables
- Powerful and quick ways to aggregate and filter data before loading it to R
- Infrastructure to store data in a relational, more structured scheme compared to the traditional matrix model of spreadsheets and R objects
- Procedures to join and merge related data
- Concurrent and network access from multiple clients at the same time
- Security policies and limits to access the data
- A scalable and configurable backend to store data
The DBI package provides a database interface, a communication channel between R and various relational database management systems (RDBMS), such as MySQL, PostgreSQL, MonetDB, Oracle, and for example Open Document Databases, and so on. There is no real need to install the package on its own because, acting as an interface, it will be installed anyway as a dependency, if needed.
Connecting to a database and fetching data is pretty similar with all these backends, as all are based on the relational model and using SQL to manage and query data. Please be advised that there are some important differences between the aforementioned database engines and that several more open-source and commercial alternatives also exist. But we will not dig into the details on how to choose a database backend or how to build a data warehouse and extract, transform, and load (ETL) workflows, but we will only concentrate on making connections and managing data from R.
Note
SQL, originally developed at IBM, with its more than 40 years of history, is one of the most important programming languages nowadays—with various dialects and implementations. Being one of the most popular declarative languages all over the world, there are many online tutorials and free courses to learn how to query and manage data with SQL, which is definitely one of the most important tools in every data scientist's Swiss army knife.
So, besides R, it's really worth knowing your way around RDBMS, which are extremely common in any industry you may be working at as a data analyst or in a similar position.
Setting up the test environment
Database backends usually run on servers remote from the users doing data analysis, but for testing purposes, it might be a good idea to install local instances on the machine running R. As the installation process can be extremely different on various operating systems, we will not enter into any details of the installation steps, but we will rather refer to where the software can be downloaded from and some further links to great resources and documentation for installation.
Please note that installing and actually trying to load data from these databases is totally optional and you do not have to follow each step—the rest of the book will not depend on any database knowledge or prior experience with databases. On the other hand, if you do not want to mess your workspace with temporary installation of multiple database applications for testing purposes, using virtual machines might be an optimal workaround. Oracle's VirtualBox provides a free and easy way of running multiple virtual machines with their dedicated operating system and userspace.
Note
For detailed instructions on how to download then import a VirtualBox image, see the Oracle section.
This way you can quickly deploy a fully functional, but disposable, database environment to test-drive the following examples of this chapter. In the following image, you can see VirtualBox with four installed virtual machines, of which three are running in the background to provide some database backends for testing purposes:

Note
VirtualBox can be installed by your operating system's package manager on Linux or by downloading the installation binary/sources from https://www.virtualbox.org/wiki/Downloads. For detailed and operating-system specific installation information, please refer to the Chapter 2, Installation details of the manual: http://www.virtualbox.org/manual/.
Nowadays, setting up and running a virtual machine is really intuitive and easy; basically you only need a virtual machine image to be loaded and launched. Some virtual machines, so called appliances, include the operating system, with a number of further software usually already configured to work, for simple, easy and quick distribution.
Tip
Once again, if you do not enjoy installing and testing new software or spending time on learning about the infrastructure empowering your data needs, the following steps are not necessary and you can freely skip these optional tasks primarily described for full-stack developers/data scientists.
Such pre-configured virtual machines to be run on any computer can be downloaded from various providers on the Internet in multiple file formats, such as OVF or OVA. General purpose VirtualBox virtual appliances can be downloaded for example from http://virtualboximages.com/vdi/index or http://virtualboxes.org/images/.
Note
Virtual appliances should be imported in VirtualBox, while non-OVF/OVA disk images should be attached to newly created virtual machines; thus, some extra manual configuration might also be needed.
Oracle also has a repository with a bunch of useful virtual images for data scientist apprentices and other developers at http://www.oracle.com/technetwork/community/developer-vm/index.html, with for example the Oracle Big Data Lite VM developer virtual appliance featuring the following most important components:
- Oracle Database
- Apache Hadoop and various tools in Cloudera distribution
- The Oracle R Distribution
- Build on Oracle Enterprise Linux
Disclaimer: Oracle wouldn't be my first choice personally, but they did a great job with their platform-independent virtualization environment, just like with providing free developer VMs based on their commercial products. In short, it's definitely worth using the provided Oracle tools.
Note
If you cannot reach your installed virtual machines on the network, please update your network settings to use Host-only adapter if no Internet connection is needed, or Bridged networking for a more robust setup. The latter setting will reserve an extra IP on your local network for the virtual machine; this way, it becomes accessible easily. Please find more details and examples with screenshots in the Oracle database section.
Another good source of virtual appliances created for open-source database engines is the Turnkey GNU/Linux repository at http://www.turnkeylinux.org/database. These images are based on Debian Linux, are totally free to use, and currently support the MySQL, PostgreSQL, MongoDB, and CouchDB databases.
A great advantage of the Turnkey Linux media is that it includes only open-source, free software and non-proprietary stuff. Besides, the disk images are a lot smaller and include only the required components for one dedicated database engine. This also results in far faster installation with less overhead in terms of the required disk and memory space.
Further similar virtual appliances are available at http://www.webuzo.com/sysapps/databases with a wider range of database backends, such as Cassandra, HBase, Neo4j, Hypertable, or Redis, although some of the Webuzo appliances might require a paid subscription for deployment.
And as the new cool being Docker, I even more suggest you to get familiar with its concept on deploying software containers incredibly fast. Such container can be described as a standalone filesystem including the operating system, libraries, tools, data and so is based on abstraction layers of Docker images. In practice this means that you can fire up a database including some demo data with a one-liner command on your localhost, and developing such custom images is similarly easy. Please see some simple examples and further references at my R and Pandoc-related Docker images described at https://github.com/cardcorp/card-rocker.
MySQL and MariaDB
MySQL is the most popular open-source database engine all over the world based on the number of mentions, job offers, Google searches, and so on, summarized by the DB-Engines Ranking: http://db-engines.com/en/ranking. Mostly used in Web development, the high popularity is probably due to the fact that MySQL is free, platform-independent, and relatively easy to set up and configure—just like its drop-in replacement fork called MariaDB.
Note
MariaDB is a community-developed, fully open-source fork of MySQL, started and led by the founder of MySQL, Michael Widenius. It was later merged with SkySQL; thus further ex-MySQL executives and investors joined the fork. MariaDB was created after Sun Microsystems bought MySQL, currently owned by Oracle, and the development of the database engine changed.
We will refer to both engines as MySQL in the book to keep it simple, as MariaDB can be considered as a drop-in replacement for MySQL, so please feel free to reproduce the following examples with either MySQL or MariaDB.
Although the installation of a MySQL server is pretty straightforward on most operating systems (https://dev.mysql.com/downloads/mysql/), one might rather prefer to have the database installed in a virtual machine. Turnkey Linux provides small but fully configured, virtual appliances for free: http://www.turnkeylinux.org/mysql.
R provides multiple ways to query data from a MySQL database. One option is to use the RMySQL package, which might be a bit tricky for some users to install. If you are on Linux, please be sure to install the development packages of MySQL along with the MySQL client, so that the package can compile on your system. And, as there are no binary packages available on CRAN for Windows installation due to the high variability of MySQL versions, Windows users should also compile the package from source:
> install.packages('RMySQL', type = 'source')
Windows users might find the following blog post useful about the detailed installation steps: http://www.ahschulz.de/2013/07/23/installing-rmysql-under-windows/.
Note
For the sake of simplicity, we will refer to the MySQL server as localhost listening on the default 3306 port; user will stand as user and password as password in all database connections. We will work with the hflights table in the hflights_db database, just like in the SQLite examples a few pages earlier. If you are working in a remote or virtual server, please modify the host, username, and so on arguments of the following code examples accordingly.
After successfully installing and starting the MySQL server, we have to set up a test database, which we could later populate in R. To this end, let us start the MySQL command-line tool to create the database and a test user.
Please note that the following example was run on Linux, and a Windows user might have to also provide the path and probably the exe file extension to start the MySQL command-line tool:

This quick session can be seen in the previous screenshot, where we first connected to the MySQL server in the command-line as the root (admin) user. Then we created a database named hflights_db, and granted all privileges and permissions of that database to a new user called user with the password set to password. Then we simply verified whether we could connect to the database with the newly created user, and we exited the command-line MySQL client.
To load data from a MySQL database into R, first we have to connect and also often authenticate with the server. This can be done with the automatically loaded
DBI package when attaching RMySQL:
> library(RMySQL) Loading required package: DBI > con <- dbConnect(dbDriver('MySQL'), + user = 'user', password = 'password', dbname = 'hflights_db')
Now we can refer to our MySQL connection as con, where we want to deploy the hflights dataset for later access:
> dbWriteTable(con, name = 'hflights', value = hflights) [1] TRUE > dbListTables(con) [1] "hflights"
The dbWriteTable function wrote the hflights data frame with the same name to the previously defined connection. The latter command shows all the tables in the currently used databases, equivalent to the SHOW TABLES SQL command. Now that we have our original CVS file imported to MySQL, let's see how long it takes to read the whole dataset:
> system.time(dbReadTable(con, 'hflights')) user system elapsed 0.993 0.000 1.058
Or we can do so with a direct SQL command passed to dbGetQuery from the same DBI package:
> system.time(dbGetQuery(con, 'select * from hflights')) user system elapsed 0.910 0.000 1.158
And, just to keep further examples simpler, let's get back to the
sqldf package, which stands for "SQL select on data frames". As a matter of fact, sqldf is a convenient wrapper around DBI's dbSendQuery function with some useful defaults, and returns data.frame. This wrapper can query various database engines, such as SQLite, MySQL, H2, or PostgreSQL, and defaults to the one specified in the global sqldf.driver option; or, if that's NULL, it will then check if any R packages have been loaded for the aforementioned backends.
As we have already loaded RMySQL, now sqldf will default to using MySQL instead of SQLite. But we still have to specify which connection to use; otherwise the function will try to open a new one—without any idea about our complex username and password combination, not to mention the mysterious database name. The connection can be passed in each sqldf expression or defined once in a global option:
> options('sqldf.connection' = con) > system.time(sqldf('select * from hflights')) user system elapsed 0.807 0.000 1.014
The difference in the preceding three versions of the same task does not seem to be significant. That 1-second timing seems to be a pretty okay result compared to our previously tested methods—although loading the whole dataset with data.table still beats this result. What about if we only need a subset of the dataset? Let's fetch only those flights ending in Nashville, just like in our previous SQLite example:
> system.time(sqldf('SELECT * FROM hflights WHERE Dest = "BNA"')) user system elapsed 0.000 0.000 0.281
This does not seem to be very convincing compared to our previous SQLite test, as the latter could reproduce the same result in less than 100 milliseconds. But please also note that that both the user and system elapsed times are zero, which was not the case with SQLite.
Note
The returned elapsed time by system.time means the number of milliseconds passed since the start of the evaluation. The user and system times are a bit trickier to understand; they are reported by the operating system. More or less, user means the CPU time spent by the called process (like R or the MySQL server), while system reports the CPU time required by the kernel and other operating system processes (such as opening a file for reading). See ?proc.time for further details.
This means that no CPU time was used at all to return the required subset of data, which took almost 100 milliseconds with SQLite. How is it possible? What if we index the database on Dest?
> dbSendQuery(con, 'CREATE INDEX Dest_idx ON hflights (Dest(3));')
This SQL query stands for creating an index named Dest_idx in our table based on the Dest column's first three letters.
Note
SQL index can seriously boost the performance of a SELECT statement with WHERE clauses, as MySQL this way does not have to read through the entire database to match each row, but it can determine the position of the relevant search results. This performance boost becomes more and more spectacular with larger databases, although it's also worth mentioning that indexing only makes sense if subsets of data are queried most of the time. If most or all data is needed, sequential reads would be faster.
Live example:
> system.time(sqldf('SELECT * FROM hflights WHERE Dest = "BNA"')) user system elapsed 0.024 0.000 0.034
It seems to be a lot better! Well, of course, we could have also indexed the SQLite database, not just the MySQL instance. To test it again, we have to revert the default sqldf driver to SQLite, which was overridden by loading the
RMySQL package:
> options(sqldf.driver = 'SQLite') > sqldf("CREATE INDEX Dest_idx ON hflights(Dest);", + dbname = "hflights_db")) NULL > system.time(sqldf("select * from hflights where + Dest = '\"BNA\"'", dbname = "hflights_db")) user system elapsed 0.034 0.004 0.036
So it seems that both database engines are capable of returning the required subset of data in a fraction of a second, which is a lot better even compared to what we achieved with the impressive data.table before.
Although SQLite proved to be faster than MySQL in some earlier examples, there are many reasons to choose the latter in most situations. First, SQLite is a file-based database, which simply means that the database should be on a filesystem attached to the computer running R. This usually means having the SQLite database and the running R session on the same computer. Similarly, MySQL can handle larger amount of data; it has user management and rule-based control on what they can do, and concurrent access to the same dataset. The smart data scientist knows how to choose his weapon—depending on the task, another database backend might be the optimal solution. Let's see what other options we have in R!
PostgreSQL
While MySQL is said to be the most popular open-source relational database management system, PostgreSQL is famous for being "the world's most advanced open source database". This means that PostgreSQL is often considered to have more features compared to the simpler but faster MySQL, including analytic functions, which has led to PostgreSQL often being described as the open-source version of Oracle.
This sounds rather funny now, as Oracle owns MySQL today. So a bunch of things have changed in the past 20-30 years of RDBMS history, and PostgreSQL is not so slow any more. On the other hand, MySQL has also gained some nice new features—for example MySQL also became ACID-compliant with the InnoDB engine, allowing rollback to previous states of the database. There are some other differences between the two popular database servers that might support choosing either of them. Now let's see what happens if our data provider has a liking for PostgreSQL instead of MySQL!
Installing PostgreSQL is similar to MySQL. One may install the software with the operating system's package manager, download a graphical installer from http://www.enterprisedb.com/products-services-training/pgdownload, or run a virtual appliance with, for example, the free Turnkey Linux, which provides a small but fully configured disk image for free at http://www.turnkeylinux.org/postgresql.
Tip
Downloading the example code
You can download the example code files from your account at http://www.packtpub.com for all the Packt Publishing books you have purchased. If you purchased this book elsewhere, you can visit http://www.packtpub.com/support and register to have the files e-mailed directly to you.
After successfully installing and starting the server, let's set up the test database—just like we did after the MySQL installation:

The syntax is a bit different in some cases, and we have used some command-line tools for the user and database creation. These helper programs are shipped with PostgreSQL by default, and MySQL also have some similar functionality with mysqladmin.
After setting up the initial test environment, or if we already have a working database instance to connect, we can repeat the previously described data management tasks with the help of the RPostgreSQL package:
> library(RPostgreSQL) Loading required package: DBI
Note
If your R session starts to throw strange error messages in the following examples, it's highly possible that the loaded R packages are conflicting. You could simply start a clean R session, or detach the previously attached packages—for example, detach('package:RMySQL', unload = TRUE).
Connecting to the database (listening on the default port number 5432) is again familiar:
> con <- dbConnect(dbDriver('PostgreSQL'), user = 'user', + password = 'password', dbname = 'hflights_db')
Let's verify that we are connected to the right database instance, which should be currently empty without the hflights table:
> dbListTables(con) character(0) > dbExistsTable(con, 'hflights') [1] FALSE
Then let's write our demo table in PostgreSQL and see if the old rumor about it being slower than MySQL is still true:
> dbWriteTable(con, 'hflights', hflights) [1] TRUE > system.time(dbReadTable(con, 'hflights')) user system elapsed 0.590 0.013 0.921
Seems to be impressive! What about loading partial data?
> system.time(dbGetQuery(con, + statement = "SELECT * FROM hflights WHERE \"Dest\" = 'BNA';")) user system elapsed 0.026 0.000 0.082
Just under 100 milliseconds without indexing! Please note the extra escaped quotes around Dest, as the default PostgreSQL behavior folds unquoted column names to lower case, which would result in a column dest does not exist error. Creating an index and running the preceding query with much improved speed can be easily reproduced based on the MySQL example.
Oracle database
Oracle Database Express Edition can be downloaded and installed from http://www.oracle.com/technetwork/database/database-technologies/express-edition/downloads/index.html. Although this is not a full-featured Oracle database, and it suffers from serious limitations, the Express Edition is a free and not too resource-hungry way to build a test environment at home.
Note
Oracle database is said to be the most popular database management system in the world, although it is available only with a proprietary license, unlike the previous two discussed RDBMSs, which means that Oracle offers the product with term licensing. On the other hand, the paid license also comes with priority support from the developer company, which is often a strict requirement in enterprise environments. Oracle Database has supported a variety of nice features since its first release in 1980, such as sharding, master-master replication, and full ACID properties.
Another way of getting a working Oracle database for testing purposes is to download an Oracle Pre-Built Developer VM from http://www.oracle.com/technetwork/community/developer-vm/index.html, or a much smaller image custom created for Hands-on Database Application Development at Oracle Technology Network Developer Day: http://www.oracle.com/technetwork/database/enterprise-edition/databaseappdev-vm-161299.html. We will follow the instructions from the latter source.
After accepting the License Agreement and registering for free at Oracle, we can download the OTN_Developer_Day_VM.ova virtual appliance. Let's import it to VirtualBox via Import appliance in the File menu, then choose the ova file, and click Next:

After clicking Import, you will have to agree again to the Software License Agreement. Importing the virtual disk image (15 GB) might take a few minutes:

After importing has finished, we should first update the networking configuration so that we can access the internal database of the virtual machine from outside. So let's switch from NAT to Bridged Adapter in the settings:

Then we can simply start the newly created virtual machine in VirtualBox. After Oracle Linux has booted, we can log in with the default oracle password.
Although we have set a bridged networking interface for our virtual machine, which means that the VM is directly connected to our real sub-network with a real IP address, the machine is not yet accessible over the network. To connect with the default DHCP settings, simply navigate to the top red bar and look for the networking icon, then select System eth0. After a few seconds the VM is accessible from your host machine, as the guest system should be connected to your network. You can verify that by running the ifconfig or ip addr show eth0 command in the already running console:

Unfortunately, this already running Oracle database is not yet accessible outside the guest machine. The developer VM comes with a rather strict firewall by default, which should be disabled first. To see the rules in effect, run the standard iptables -L -n command and, to flush all rules, execute iptables -F:

Now that we have a running and remotely accessible Oracle database, let's prepare the R client side. Installing the ROracle package might get tricky on some operating systems, as there are no prebuilt binary packages and you have to manually install the Oracle Instant Client Lite and SDK libraries before compiling the package from source. If the compiler complained about the path of your previously installed Oracle libraries, please pass the --with-oci-lib and --with-oci-inc arguments with your custom paths with the --configure-args parameter. More details can be found in the package installation document: http://cran.r-project.org/web/packages/ROracle/INSTALL.
For example, on Arch Linux you can install the Oracle libs from AUR, then run the following command in bash after downloading the R package from CRAN:
# R CMD INSTALL --configure-args='--with-oci-lib=/usr/include/ \ > --with-oci-inc=/usr/share/licenses/oracle-instantclient-basic' \ > ROracle_1.1-11.tar.gz
After installing and loading the package, opening a connection is extremely similar to the pervious examples with DBI::dbConnect. We only pass an extra parameter here. First, let us specify the hostname or direct IP address of the Oracle database included in the dbname argument. Then we can connect to the already existing PDB1 database of the developer machine instead of the previously used hflights_db—just to save some time and space in the book on slightly off-topic database management tasks:
> library(ROracle) Loading required package: DBI > con <- dbConnect(dbDriver('Oracle'), user = 'pmuser', + password = 'oracle', dbname = '//192.168.0.16:1521/PDB1')
And we have a working connection to Oracle RDBMS:
> summary(con) User name: pmuser Connect string: //192.168.0.16:1521/PDB1 Server version: 12.1.0.1.0 Server type: Oracle RDBMS Results processed: 0 OCI prefetch: FALSE Bulk read: 1000 Statement cache size: 0 Open results: 0
Let's see what we have in the bundled database on the development VM:
> dbListTables(con) [1] "TICKER_G" "TICKER_O" "TICKER_A" "TICKER"
So it seems that we have a table called TICKER with three views on tick data of three symbols. Saving the hflights table in the same database will not do any harm, and we can also instantly test the speed of the Oracle database when reading the whole table:
> dbWriteTable(con, 'hflights', hflights) [1] TRUE > system.time(dbReadTable(con, 'hflights')) user system elapsed 0.980 0.057 1.256
And the extremely familiar subset with 3,481 cases:
> system.time(dbGetQuery(con, + "SELECT * FROM \"hflights\" WHERE \"Dest\" = 'BNA'")) user system elapsed 0.046 0.003 0.131
Please note the quotes around the table name. In the previous examples with MySQL and PostgreSQL, the SQL statements run fine without those. However, the quotes are needed in the Oracle database, as we have saved the table with an all-lowercase name, and the default rule in Oracle DB is to store object names in upper case. The only other option is to use double quotes to create them, which is what we did; thus we have to refer to the table with quotes around the lowercase name.
Note
We started with unquoted table and column names in MySQL, then had to add escaped quotes around the variable name in the PostgreSQL query run from R, and now in Oracle database we have to put both names between quotes—which demonstrates the slight differences in the various SQL flavors (such as MySQL, PostgreSQL, PL/SQL of Oracle or Microsoft's Transact-SQL) on top of ANSI SQL.
And more importantly: do not stick to one database engine with all your projects, but rather choose the optimal DB for the task if company policy doesn't stop you doing so.
These results were not so impressive compared to what we have seen by PostgreSQL, so let's also see the results of an indexed query:
> dbSendQuery(con, 'CREATE INDEX Dest_idx ON "hflights" ("Dest")') Statement: CREATE INDEX Dest_idx ON "hflights" ("Dest") Rows affected: 0 Row count: 0 Select statement: FALSE Statement completed: TRUE OCI prefetch: FALSE Bulk read: 1000 > system.time(dbGetQuery(con, "SELECT * FROM \"hflights\" + WHERE \"Dest\" = 'BNA'")) user system elapsed 0.023 0.000 0.069
I leave the full-scale comparative testing and benchmarking to you, so that you can run custom queries in the tests fitting your exact needs. It is highly possible that the different database engines perform differently in special use cases.
To make this process a bit more seamless and easier to implement, let's check out another R way of connecting to databases, although probably with a slight performance trade-off. For a quick scalability and performance comparison on connecting to Oracle databases with different approaches in R, please see https://blogs.oracle.com/R/entry/r_to_oracle_database_connectivity.
ODBC database access
As mentioned earlier, installing the native client software, libraries, and header files for the different databases so that the custom R packages can be built from source can be tedious and rather tricky in some cases. Fortunately, we can also try to do the opposite of this process. An alternative solution can be installing a middleware Application Programming Interface (API) in the databases, so that R, or as a matter of fact any other tool, could communicate with them in a standardized and more convenient way. However, please be advised that this more convenient way impairs performance due to the translation layer between the application and the DBMS.
The RODBC package implements access to such a layer. The Open Database Connectivity (ODBC) driver is available for most database management systems, even for CSV and Excel files, so RODBC provides a standardized way to access data in almost any databases if the ODBC driver is installed. This platform-independent interface is available for SQLite, MySQL, MariaDB, PostgreSQL, Oracle database, Microsoft SQL Server, Microsoft Access, and IBM DB2 on Windows and on Linux.
For a quick example, let's connect to MySQL running on localhost (or on a virtual machine). First, we have to set up a
Database Source Name (DSN) with the connection details, such as:
- Database driver
- Host name or address and port number, optionally a Unix socket
- Database name
- Optionally the username and password to be used for the connection
This can be done in the command line by editing the odbc.ini and odbcinst.ini files on Linux after installing the
unixODBC program. The latter should include the following configuration for the MySQL driver in your /etc folder:
[MySQL] Description = ODBC Driver for MySQL Driver = /usr/lib/libmyodbc.so Setup = /usr/lib/libodbcmyS.so FileUsage = 1
The odbc.ini file includes the aforementioned DSN configuration for the exact database and server:
[hflights] Description = MySQL hflights test Driver = MySQL Server = localhost Database = hflights_db Port = 3306 Socket = /var/run/mysqld/mysqld.sock
Or use a graphical user interface on Mac OS or Windows, as shown in the following screenshot:

After configuring a DSN, we can connect with a one-line command:
> library(RODBC) > con <- odbcConnect("hflights", uid = "user", pwd = "password")
Let's fetch the data we saved in the database before:
> system.time(hflights <- sqlQuery(con, "select * from hflights")) user system elapsed 3.180 0.000 3.398
Well, it took a few seconds to finish. That's the trade-off for using a more convenient and high-level interface to interact with the database. Removing and uploading data to the database can be done with similar high-level functions (such as sqlFetch) besides the odbc* functions, providing low-level access to the database. Quick examples:
> sqlDrop(con, 'hflights') > sqlSave(con, hflights, 'hflights')
You can use the exact same commands to query any of the other supported database engines; just be sure to set up the DSN for each backend, and to close your connections if not needed any more:
> close(con)
The RJDBC package can provide a similar interface to database management systems with a Java Database Connectivity (JDBC) driver.
Using a graphical user interface to connect to databases
Speaking of high-level interfaces, R also has a graphical user interface to connect to MySQL in the dbConnect package:
> library(dbConnect) Loading required package: RMySQL Loading required package: DBI Loading required package: gWidgets > DatabaseConnect() Loading required package: gWidgetsRGtk2 Loading required package: RGtk2
No parameters, no custom configuration in the console, just a simple dialog window:

After providing the required connection information, we can easily view the raw data and the column/variable types, and run custom SQL queries. A basic query builder can also help novice users to fetch subsamples from the database:
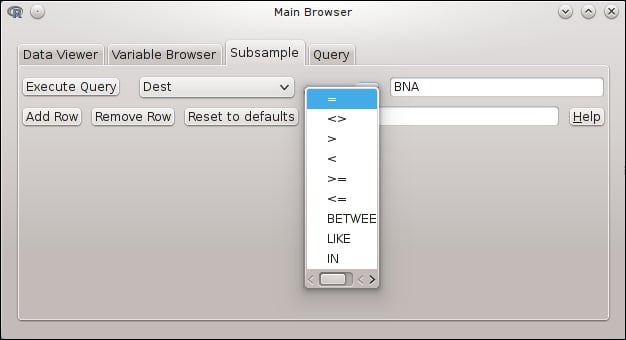
The package ships with a handy function called sqlToR, which can turn the SQL results into R objects with a click in the GUI. Unfortunately, dbConnect relies heavily on RMySQL, which means it's a MySQL-only package, and there is no plan to extend the functionality of this interface.
Other database backends
Besides the previously mentioned popular databases, there are several other implementations that we cannot discuss here in detail.
For example, column-oriented database management systems, such as MonetDB, are often used to store large datasets with millions of rows and thousands of columns to provide the backend for high-performance data mining. It also has great R support with the MonetDB.R package, which was among the most exciting talks at the useR! 2013 conference.
The ever-growing popularity of the NoSQL ecosystem also provides similar approaches, although usually without supporting SQL and providing a schema-free data storage. Apache Cassandra is a good example of such a similar, column-oriented, and primarily distributed database management system with high availably and performance, run on commodity hardware. The
RCassandra package provides access to the basic Cassandra features and the Cassandra Query Language in a convenient way with the RC.* function family. Another Google Bigtable-inspired and similar database engine is HBase, which is supported by the
rhbase package, part of the RHadoop project: https://github.com/RevolutionAnalytics/RHadoop/wiki.
Speaking of Massively Parallel Processing, HP's Vertica and Cloudera's open-source Impala are also accessible from R, so you can easily access and query large amount of data with relatively good performance.
One of the most popular NoSQL databases is MongoDB, which provides document-oriented data storage in a JSON-like format, providing an infrastructure to dynamic schemas. MongoDB is actively developed and has some SQL-like features, such as a query language and indexing, also with multiple R packages providing access to this backend. The
RMongo package uses the mongo-java-driver and thus depends on Java, but provides a rather high-level interface to the database. Another implementation, the rmongodb package, is developed and maintained by the MongoDB Team. The latter has more frequent updates and more detailed documentation, but the R integration seems to be a lot more seamless with the first package as rmongodb provides access to the raw MongoDB functions and BSON objects, instead of concentrating on a translation layer for general R users. A more recent and really promising package supporting MongoDB is mongolite developed by Jeroen Ooms.
CouchDB, my personal favorite for most schema-less projects, provides very convenient document storage with JSON objects and HTTP API, which means that integrating in applications, such as any R script, is really easy with, for example, the
RCurl package, although you may find the R4CouchDB more quick to act in interacting with the database.
Google BigQuery also provides a similar, REST-based HTTP API to query even terabytes of data hosted in the Google infrastructure with an SQL-like language. Although the
bigrquery package is not available on CRAN yet, you may easily install it from GitHub with the devtools package from the same author, Hadley Wickham:
> library(devtools) > install_github('bigrquery', 'hadley')
To test-drive the features of this package and Google BigQuery, you can sign up for a free account to fetch and process the demo dataset provided by Google, respecting the 10,000 requests per day limitation for free usage. Please note that the current implementation is a read-only interface to the database.
For rather similar database engines and comparisons, see for example http://db-engines.com/en/systems. Most of the popular databases already have R support but, if not, I am pretty sure that someone is already working on it. It's worth checking the CRAN packages at http://cran.r-project.org/web/packages/available_packages_by_name.html or searching on GitHub or on http://R-bloggers.com to see how other R users manage to interact with your database of choice.






















































