Spectral Clustering is a more sophisticated approach based on the G={V, E} graph of the dataset. The set of vertices, V, is made up of the samples, while the edges, E, connecting two different samples are weighted according to an affinity measure, whose value is proportional to the distance of two samples in the original space or in a more suitable one (in a way analogous to Kernel SVMs).
If there are n samples, it's helpful to introduce a symmetric affinity matrix:

Each element wij represents a measure of affinity between two samples. The most diffuse measures (also supported by scikit-learn) are the Radial Basis Function (RBF) and k-Nearest Neighbors (k-NN). The former is defined as follows:

The latter is based on a parameter, k, defining the number of neighbors:
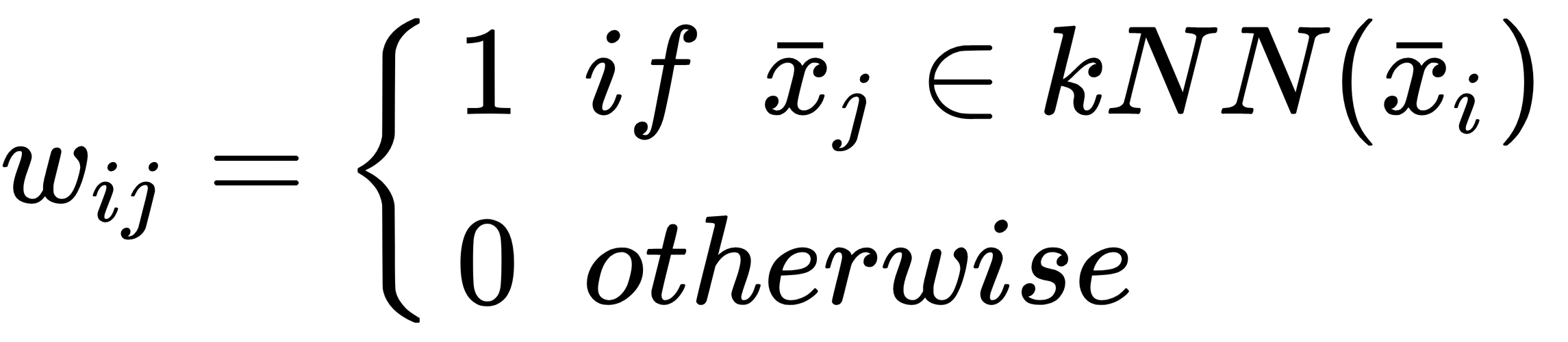
RBF is always non-null, while k-NN can yield singular affinity matrices if the graph...


































































