The next steps are specific to the PyCharm IDE. To configure DeepChem with PyCharm, we have to focus on our Conda-based installation that we did earlier in the previous section. Assuming the Conda environment is created, you will already have a DeepChem development environment:
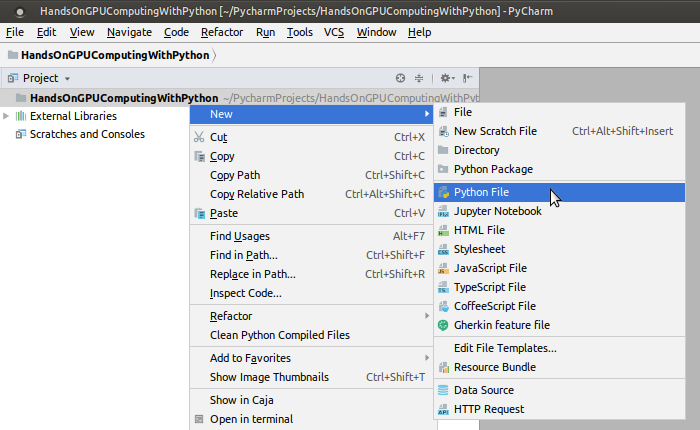
Now, you can import deepchem within your Python programs. As you can see, PyCharm Edu detects and recommends this as you begin to type import deepchem:

At this stage, we are done configuring DeepChem on the PyCharm IDE for a hands-on molecular deep learning experience with GPUs.



































































