The MNIST dataset is included in keras and can be accessed using the dataset_mnist() function:
- Let's load the data into the R environment:
mnist <- dataset_mnist()
x_train <- mnist$train$x
y_train <- mnist$train$y
x_test <- mnist$test$x
y_test <- mnist$test$y
- Our training data is of the form (images, width, height). Due to this, we'll convert the data into a one-dimensional array and rescale it:
# Reshaping the data
x_train <- array_reshape(x_train , c(nrow(x_train),784))
x_test <- array_reshape(x_test , c(nrow(x_test),784))
# Rescaling the data
x_train <- x_train/255
x_test <- x_test/255
- Our target data is an integer vector and contains values from 0 to 9. We need to one-hot encode our target variable in order to convert it into a binary matrix format. We use the to_categorical() function from keras to do this:
y_train <- to_categorical(y_train,10)
y_test <- to_categorical(y_test,10)
- Now, we can build the model. We use the Sequential API from keras to configure this model. Note that in the first layer's configuration, the input_shape argument is the shape of the input data; that is, it's a numeric vector of length 784 and represents a grayscale image. The final layer outputs a length 10 numeric vector (probabilities for each digit from 0 to 9) using a softmax activation function:
model <- keras_model_sequential()
model %>%
layer_dense(units = 256, activation = 'relu', input_shape = c(784)) %>%
layer_dropout(rate = 0.4) %>%
layer_dense(units = 128, activation = 'relu') %>%
layer_dropout(rate = 0.3) %>%
layer_dense(units = 10, activation = 'softmax')
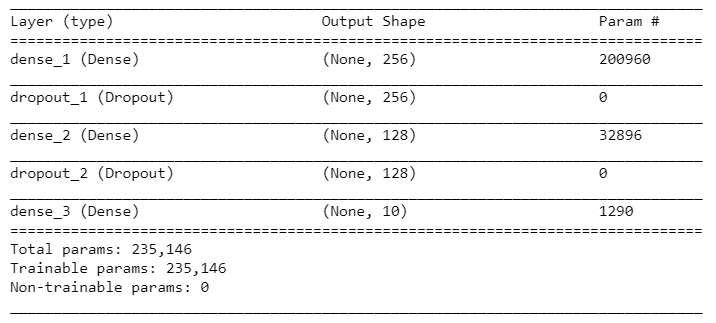
Let's look at the details of the model:
summary(model)
Here's the model's summary:
- Next, we go ahead and compile our model by providing some appropriate arguments, such as the loss function, optimizer, and metrics. Here, we have used the rmsprop optimizer. This optimizer is similar to the gradient descent optimizer, except that it can increase our learning rate so that our algorithm can take larger steps in the horizontal direction, thus converging faster:
model %>% compile(
loss = 'categorical_crossentropy',
optimizer = optimizer_rmsprop(),
metrics = c('accuracy')
)
- Now, let's fit the training data to the configured model. Here, we've set the number of epochs to 30, the batch size to 128, and the validation percentage to 20:
history <- model %>% fit(
x_train, y_train,
epochs = 30, batch_size = 128,
validation_split = 0.2
)
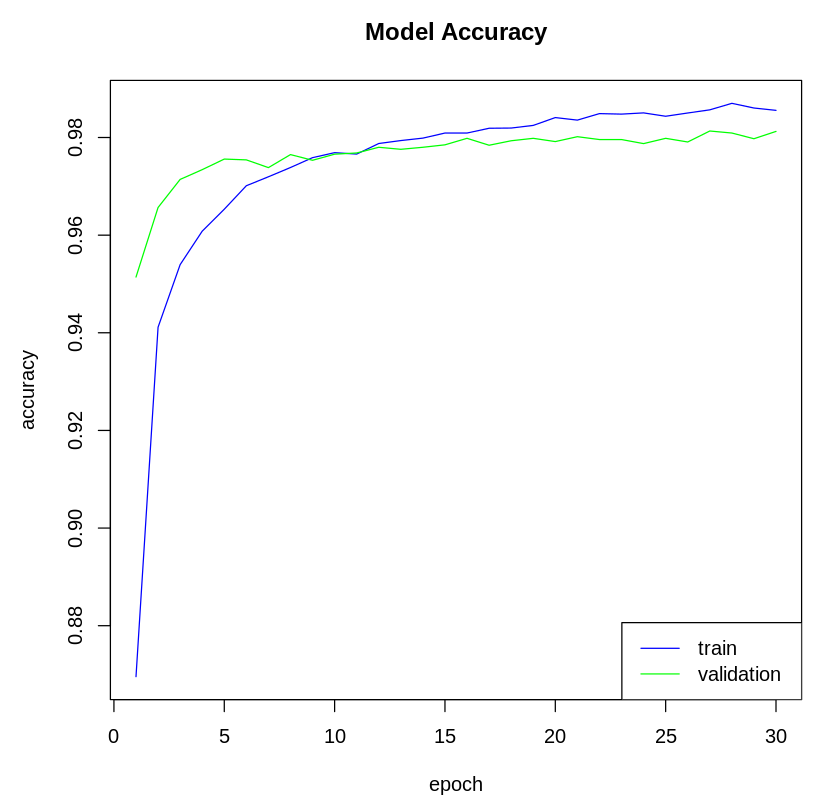
- Next, we visualize the model metrics. We can plot the model's accuracy and loss metrics from the history variable. Let's plot the model's accuracy:
# Plot the accuracy of the training data
plot(history$metrics$acc, main="Model Accuracy", xlab = "epoch", ylab="accuracy", col="blue",
type="l")
# Plot the accuracy of the validation data
lines(history$metrics$val_acc, col="green")
# Add Legend
legend("bottomright", c("train","validation"), col=c("blue", "green"), lty=c(1,1))
The following plot shows the model's accuracy on the training and test dataset:
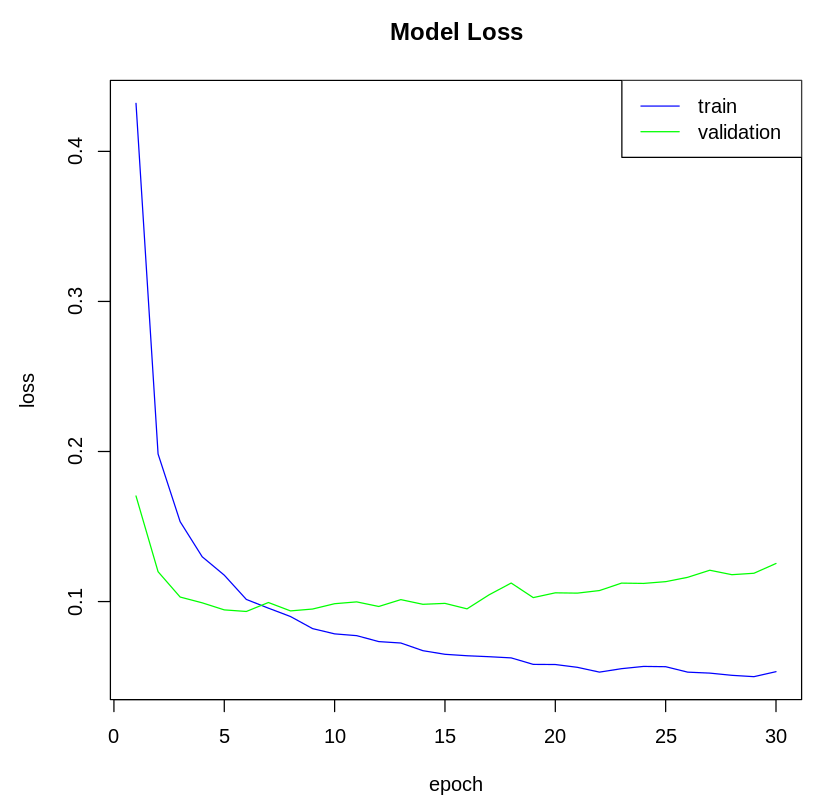
Now, let's plot the model's loss:
# Plot the model loss of the training data
plot(history$metrics$loss, main="Model Loss", xlab = "epoch", ylab="loss", col="blue", type="l")
# Plot the model loss of the validation data
lines(history$metrics$val_loss, col="green")
# Add legend
legend("topright", c("train","validation"), col=c("blue", "green"), lty=c(1,1))
The following plot shows the model's loss on the training and test dataset:
- Now, we predict the classes for the test data instances using the trained model:
model %>% predict_classes(x_test)
- Let's check the accuracy of the model on the test data:
model %>% evaluate(x_test, y_test)
The following diagram shows the model metrics on the test data:
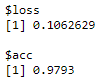
Here, we got an accuracy of around 97.9 %.

















































































